215 lines
7.6 KiB
Markdown
215 lines
7.6 KiB
Markdown
|
|
# Mental Health in the EA Community using SSC's 2019 Survey
|
|||
|
|
|
|||
|
|
If you naïvely run some regressions, you get a significant correlation between EA affiliation and mental conditions. If you look at the plots, this seems like maybe an artifact of different proportions of NAs.
|
|||
|
|
|
|||
|
|
Anyways, the numerical results are:
|
|||
|
|
|
|||
|
|
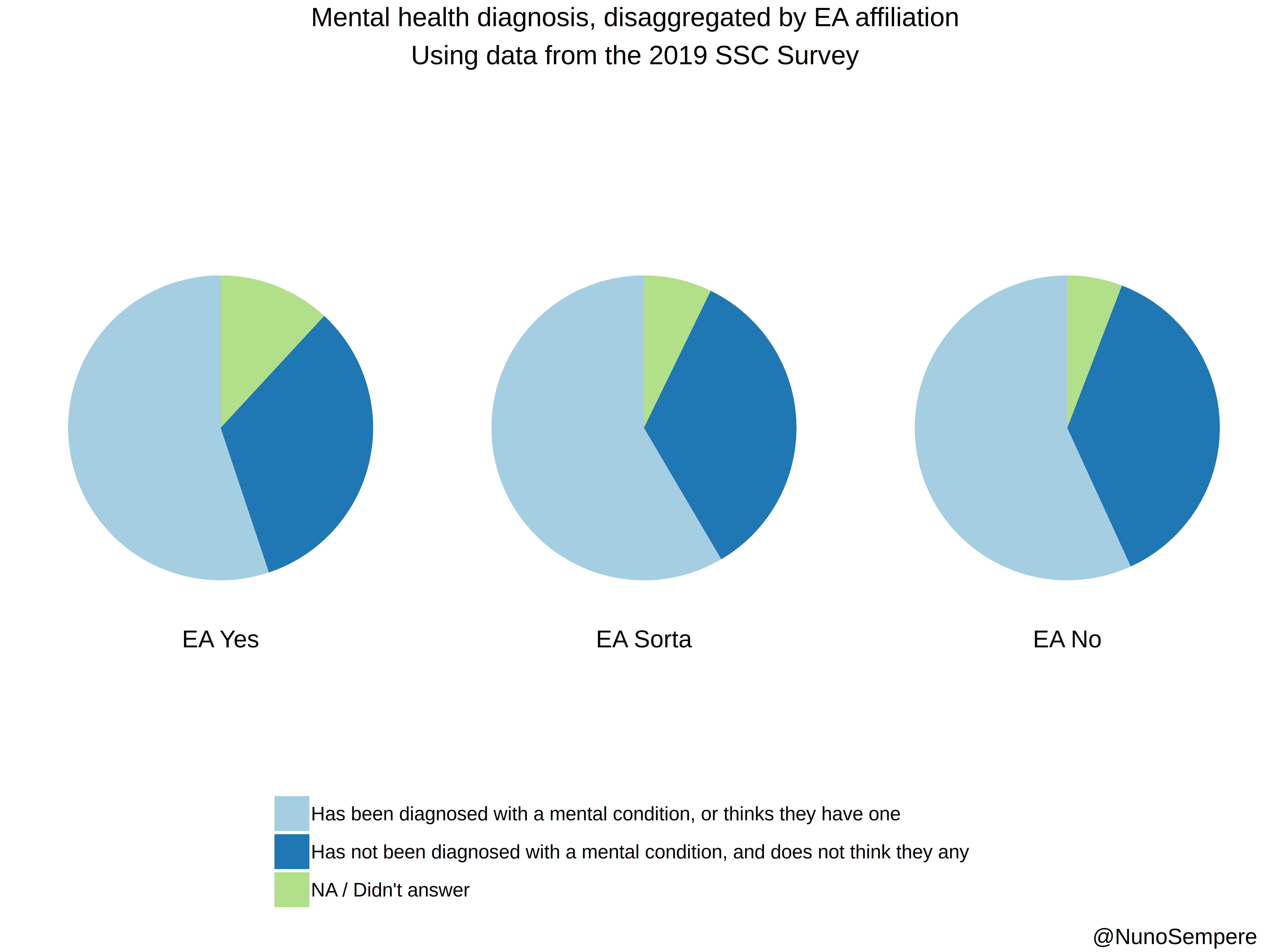
## Diagnosed + Intuited
|
|||
|
|
|
|||
|
|
```
|
|||
|
|
x y %
|
|||
|
|
1 EA Yes 959 100.00000
|
|||
|
|
2 Has been diagnosed with a mental condition, or thinks they have one 580 60.47967
|
|||
|
|
3 Has not been diagnosed with a mental condition, and does not think they any 347 36.18352
|
|||
|
|
4 NA / Didn't answer 125 13.03441
|
|||
|
|
```
|
|||
|
|
```
|
|||
|
|
x y %
|
|||
|
|
1 EA Sorta 2223 100.000000
|
|||
|
|
2 Has been diagnosed with a mental condition, or thinks they have one 1354 60.908682
|
|||
|
|
3 Has not been diagnosed with a mental condition, and does not think they any 795 35.762483
|
|||
|
|
4 NA / Didn't answer 167 7.512371
|
|||
|
|
```
|
|||
|
|
|
|||
|
|
```
|
|||
|
|
x y %
|
|||
|
|
1 EA No 4158 100.000000
|
|||
|
|
2 Has been diagnosed with a mental condition, or thinks they have one 2416 58.104858
|
|||
|
|
3 Has not been diagnosed with a mental condition, and does not think they any 1587 38.167388
|
|||
|
|
4 NA / Didn't answer 248 5.964406
|
|||
|
|
```
|
|||
|
|
|
|||
|
|
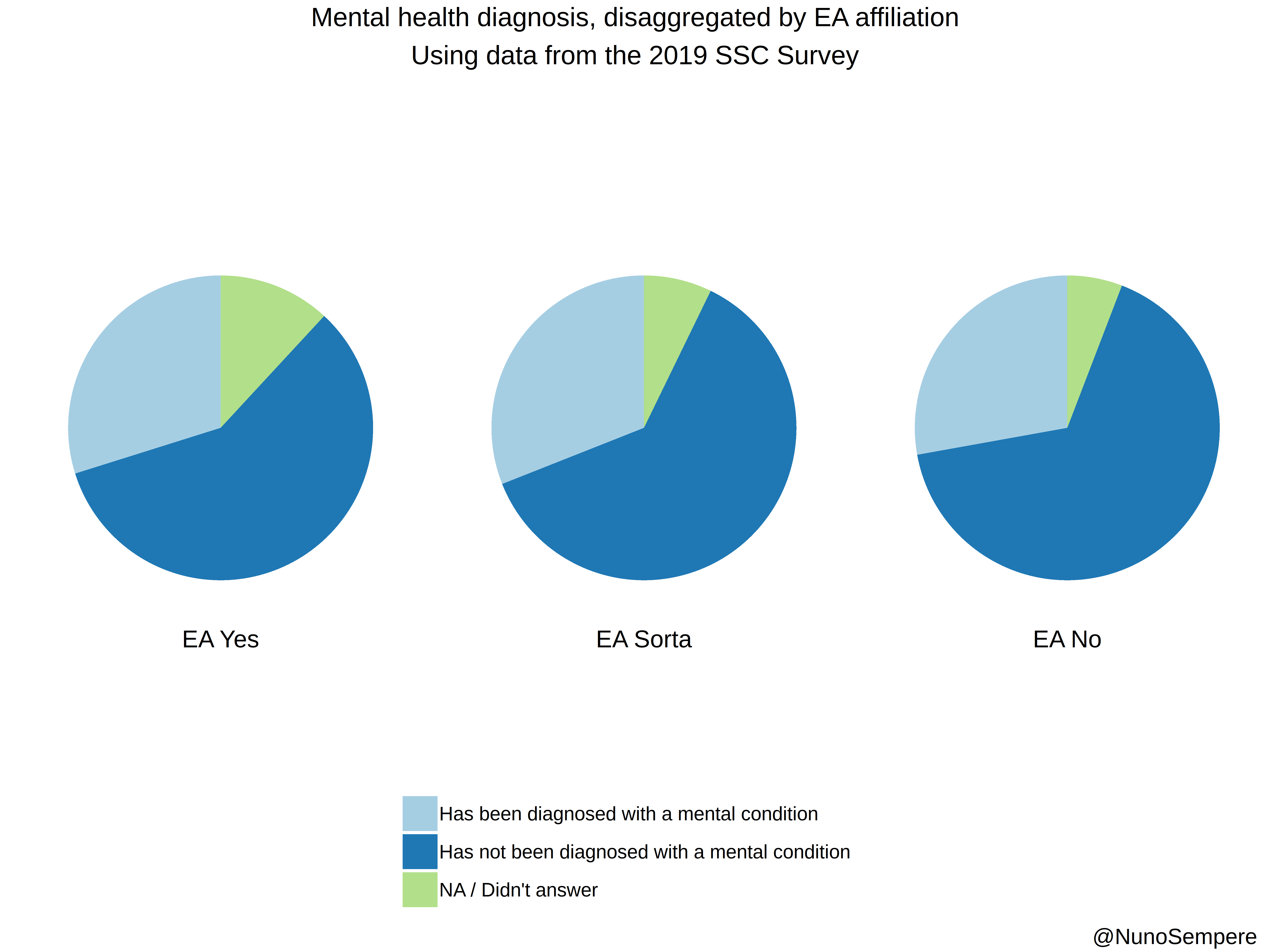
## Diagnosed
|
|||
|
|
```
|
|||
|
|
x y %
|
|||
|
|
1 EA Yes 959 100.00000
|
|||
|
|
2 Has been diagnosed with a mental condition 314 32.74244
|
|||
|
|
3 Has not been diagnosed with a mental condition 613 63.92075
|
|||
|
|
4 NA / Didn't answer 125 13.03441
|
|||
|
|
```
|
|||
|
|
```
|
|||
|
|
x y %
|
|||
|
|
1 EA Sorta 2223 100.000000
|
|||
|
|
2 Has been diagnosed with a mental condition 718 32.298695
|
|||
|
|
3 Has not been diagnosed with a mental condition 1431 64.372470
|
|||
|
|
4 NA / Didn't answer 167 7.512371
|
|||
|
|
```
|
|||
|
|
```
|
|||
|
|
x y %
|
|||
|
|
1 EA No 4158 100.000000
|
|||
|
|
2 Has been diagnosed with a mental condition 1183 28.451178
|
|||
|
|
3 Has not been diagnosed with a mental condition 2820 67.821068
|
|||
|
|
4 NA / Didn't answer 248 5.964406
|
|||
|
|
```
|
|||
|
|
|
|||
|
|
## Regressions
|
|||
|
|
### Linear
|
|||
|
|
```
|
|||
|
|
> # D$mentally_ill = Number of diagnosed mental ilnesses
|
|||
|
|
> # D$mentally_ill2= Number of mental ilnesses, diagnosed + intuited
|
|||
|
|
```
|
|||
|
|
```
|
|||
|
|
> summary(lm(D$mentally_ill ~ D$`EA ID`))
|
|||
|
|
|
|||
|
|
Call:
|
|||
|
|
lm(formula = D$mentally_ill ~ D$`EA ID`)
|
|||
|
|
|
|||
|
|
Residuals:
|
|||
|
|
Min 1Q Median 3Q Max
|
|||
|
|
-0.5717 -0.5514 -0.4689 0.4486 10.4283
|
|||
|
|
|
|||
|
|
Coefficients:
|
|||
|
|
Estimate Std. Error t value Pr(>|t|)
|
|||
|
|
(Intercept) 0.46890 0.01424 32.935 < 2e-16 ***
|
|||
|
|
D$`EA ID`Sorta 0.08252 0.02409 3.426 0.000617 ***
|
|||
|
|
D$`EA ID`Yes 0.10284 0.03283 3.132 0.001742 **
|
|||
|
|
---
|
|||
|
|
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
|
|||
|
|
|
|||
|
|
Residual standard error: 0.9008 on 7076 degrees of freedom
|
|||
|
|
(354 observations deleted due to missingness)
|
|||
|
|
Multiple R-squared: 0.002421, Adjusted R-squared: 0.002139
|
|||
|
|
F-statistic: 8.587 on 2 and 7076 DF, p-value: 0.0001884
|
|||
|
|
```
|
|||
|
|
```
|
|||
|
|
> summary(lm(D$mentally_ill2 ~ D$`EA ID`))
|
|||
|
|
|
|||
|
|
Call:
|
|||
|
|
lm(formula = D$mentally_ill2 ~ D$`EA ID`)
|
|||
|
|
|
|||
|
|
Residuals:
|
|||
|
|
Min 1Q Median 3Q Max
|
|||
|
|
-1.3711 -1.2638 -0.2638 0.7362 9.6289
|
|||
|
|
|
|||
|
|
Coefficients:
|
|||
|
|
Estimate Std. Error t value Pr(>|t|)
|
|||
|
|
(Intercept) 1.26380 0.02243 56.343 <2e-16 ***
|
|||
|
|
D$`EA ID`Sorta 0.09637 0.03795 2.539 0.0111 *
|
|||
|
|
D$`EA ID`Yes 0.10729 0.05173 2.074 0.0381 *
|
|||
|
|
---
|
|||
|
|
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
|
|||
|
|
|
|||
|
|
Residual standard error: 1.419 on 7076 degrees of freedom
|
|||
|
|
(354 observations deleted due to missingness)
|
|||
|
|
Multiple R-squared: 0.001216, Adjusted R-squared: 0.0009338
|
|||
|
|
F-statistic: 4.308 on 2 and 7076 DF, p-value: 0.0135
|
|||
|
|
```
|
|||
|
|
```
|
|||
|
|
> summary(lm(D$mentally_ill>0 ~ D$`EA ID`))
|
|||
|
|
|
|||
|
|
Call:
|
|||
|
|
lm(formula = D$mentally_ill > 0 ~ D$`EA ID`)
|
|||
|
|
|
|||
|
|
Residuals:
|
|||
|
|
Min 1Q Median 3Q Max
|
|||
|
|
-0.3387 -0.3341 -0.2955 0.6659 0.7045
|
|||
|
|
|
|||
|
|
Coefficients:
|
|||
|
|
Estimate Std. Error t value Pr(>|t|)
|
|||
|
|
(Intercept) 0.295528 0.007323 40.354 < 2e-16 ***
|
|||
|
|
D$`EA ID`Sorta 0.038581 0.012391 3.114 0.00186 **
|
|||
|
|
D$`EA ID`Yes 0.043199 0.016889 2.558 0.01055 *
|
|||
|
|
---
|
|||
|
|
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
|
|||
|
|
|
|||
|
|
Residual standard error: 0.4633 on 7076 degrees of freedom
|
|||
|
|
(354 observations deleted due to missingness)
|
|||
|
|
Multiple R-squared: 0.001835, Adjusted R-squared: 0.001553
|
|||
|
|
F-statistic: 6.505 on 2 and 7076 DF, p-value: 0.001505
|
|||
|
|
```
|
|||
|
|
```
|
|||
|
|
> summary(lm(D$mentally_ill2>0 ~ D$`EA ID`))
|
|||
|
|
|
|||
|
|
Call:
|
|||
|
|
lm(formula = D$mentally_ill2 > 0 ~ D$`EA ID`)
|
|||
|
|
|
|||
|
|
Residuals:
|
|||
|
|
Min 1Q Median 3Q Max
|
|||
|
|
-0.6301 -0.6036 0.3699 0.3965 0.3965
|
|||
|
|
|
|||
|
|
Coefficients:
|
|||
|
|
Estimate Std. Error t value Pr(>|t|)
|
|||
|
|
(Intercept) 0.603547 0.007692 78.466 <2e-16 ***
|
|||
|
|
D$`EA ID`Sorta 0.026513 0.013014 2.037 0.0417 *
|
|||
|
|
D$`EA ID`Yes 0.022127 0.017738 1.247 0.2123
|
|||
|
|
---
|
|||
|
|
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
|
|||
|
|
|
|||
|
|
Residual standard error: 0.4867 on 7076 degrees of freedom
|
|||
|
|
(354 observations deleted due to missingness)
|
|||
|
|
Multiple R-squared: 0.0006657, Adjusted R-squared: 0.0003832
|
|||
|
|
F-statistic: 2.357 on 2 and 7076 DF, p-value: 0.09481
|
|||
|
|
```
|
|||
|
|
|
|||
|
|
## Logistic
|
|||
|
|
|
|||
|
|
```
|
|||
|
|
> summary(glm(D$mentally_ill>0 ~ D$`EA ID`, family=binomial(link='logit')))
|
|||
|
|
|
|||
|
|
Call:
|
|||
|
|
glm(formula = D$mentally_ill > 0 ~ D$`EA ID`, family = binomial(link = "logit"))
|
|||
|
|
|
|||
|
|
Deviance Residuals:
|
|||
|
|
Min 1Q Median 3Q Max
|
|||
|
|
-0.9095 -0.9018 -0.8370 1.4807 1.5614
|
|||
|
|
|
|||
|
|
Coefficients:
|
|||
|
|
Estimate Std. Error z value Pr(>|z|)
|
|||
|
|
(Intercept) -0.86868 0.03464 -25.078 < 2e-16 ***
|
|||
|
|
D$`EA ID`Sorta 0.17902 0.05737 3.120 0.00181 **
|
|||
|
|
D$`EA ID`Yes 0.19971 0.07756 2.575 0.01003 *
|
|||
|
|
---
|
|||
|
|
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
|
|||
|
|
|
|||
|
|
(Dispersion parameter for binomial family taken to be 1)
|
|||
|
|
|
|||
|
|
Null deviance: 8797.8 on 7078 degrees of freedom
|
|||
|
|
Residual deviance: 8784.8 on 7076 degrees of freedom
|
|||
|
|
(354 observations deleted due to missingness)
|
|||
|
|
AIC: 8790.8
|
|||
|
|
|
|||
|
|
Number of Fisher Scoring iterations: 4
|
|||
|
|
```
|
|||
|
|
```
|
|||
|
|
> summary(glm(D$mentally_ill2>0 ~ D$`EA ID`, family=binomial(link='logit')))
|
|||
|
|
|
|||
|
|
Call:
|
|||
|
|
glm(formula = D$mentally_ill2 > 0 ~ D$`EA ID`, family = binomial(link = "logit"))
|
|||
|
|
|
|||
|
|
Deviance Residuals:
|
|||
|
|
Min 1Q Median 3Q Max
|
|||
|
|
-1.4103 -1.3603 0.9612 1.0049 1.0049
|
|||
|
|
|
|||
|
|
Coefficients:
|
|||
|
|
Estimate Std. Error z value Pr(>|z|)
|
|||
|
|
(Intercept) 0.42027 0.03231 13.007 <2e-16 ***
|
|||
|
|
D$`EA ID`Sorta 0.11221 0.05514 2.035 0.0419 *
|
|||
|
|
D$`EA ID`Yes 0.09344 0.07517 1.243 0.2139
|
|||
|
|
---
|
|||
|
|
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
|
|||
|
|
|
|||
|
|
(Dispersion parameter for binomial family taken to be 1)
|
|||
|
|
|
|||
|
|
Null deviance: 9439.1 on 7078 degrees of freedom
|
|||
|
|
Residual deviance: 9434.4 on 7076 degrees of freedom
|
|||
|
|
(354 observations deleted due to missingness)
|
|||
|
|
AIC: 9440.4
|
|||
|
|
|
|||
|
|
Number of Fisher Scoring iterations: 4
|
|||
|
|
```
|
|||
|
|
|
|||
|
|
## Plots:
|
|||
|
|
|
|||
|
|
|
|||
|
|
|